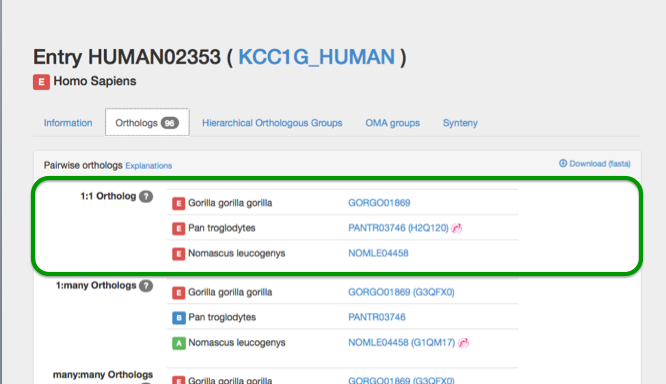
Types of orthologs
Homologous sequences are sequence that sharing a common ancestry, be they within or between species. They can be further classified in two main categories: orthologs, which are pairs of genes that started diverging through speciation, and paralogs, which are pairs of genes that started diverging through gene duplication.
The OMA Browser reports different subtypes of orthologous and paralogous relationships:
The entry has only one ortholog in the other species and the orthologous entries have only one ortholog in this species.
The entry has more than one ortholog in the other species but all orthologous entries have only one ortholog in this species. This implies that the gene was duplicated in an ancestor of the other species, but after the speciation event.
The entry has only one ortholog in the other species, but the orthologous entry has more than one ortholog in this species. This implies that the gene was duplicated in an ancestor of this species, but after the speciation event.
The entry has more than one ortholog in the other species and the orthologous entries have more than one ortholog in this species. This implies that the gene was duplicated at least twice: in the lineage of the query species and in the lineage of the other species, yet all descend from a common ancestral gene in the last common ancestor of the two species.
We call "close" paralogs the paralogous sequence pairs that are co-orthologous to at least one other entry reported in the list of orthologs. Or in other words, they are in-paralogs with respect to the last common ancestor of all the species represented among the orthologs.
Hierarchical groups contain genes that descend from a single common ancestral gene within a given taxonomic range.
OMA groups contain sets of genes which are all orthologous to one another within group. This implies that there is at most one entry from each species in a group.